Frequently Asked Questions
To inquire/order services, please review this guide and completely fill in its entirely the ordering forms related to your project for accurate processing. Email the completed form to
- Sample Submission for DNA Projects Download PDF
- Sample Submission for Tissue (to be used for DNA isolation) Download PDF
- Sample Submission for tGBS® Genotyping-by-Sequencing (GBS) Projects Download PDF
Sample Submission for DNA Projects
Sample Requirements
At least 5 µgrams of double stranded DNA isolated using a column or gel purification protocol with a 260/280 ratio of at least 1.8. If sufficient DNA is available, we recommend 10 µgrams. As mentioned above, for dried samples we recommend starting with 20 µgrams of total DNA to compensate for sample loss and degradation during drying and re-elution.
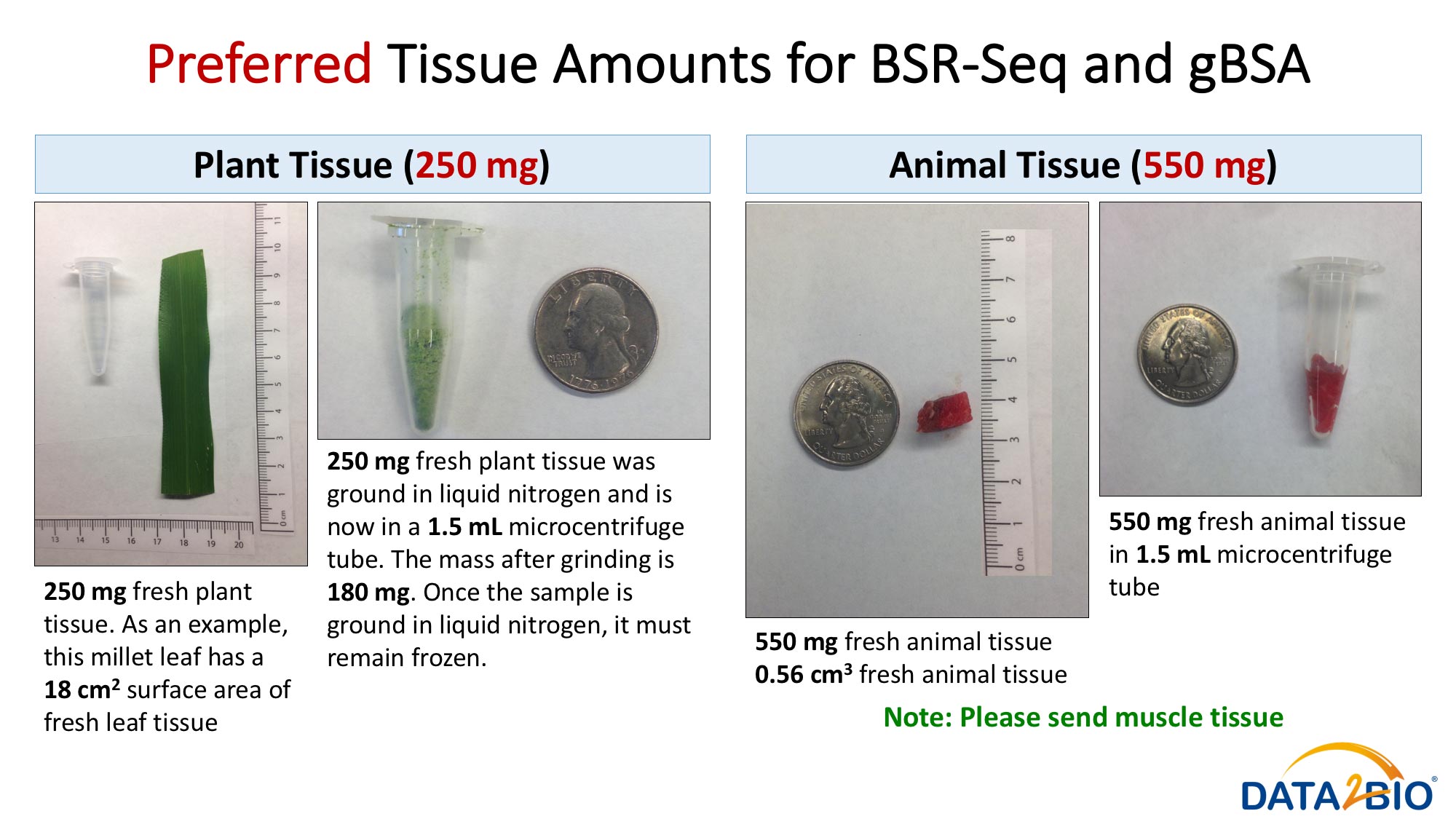
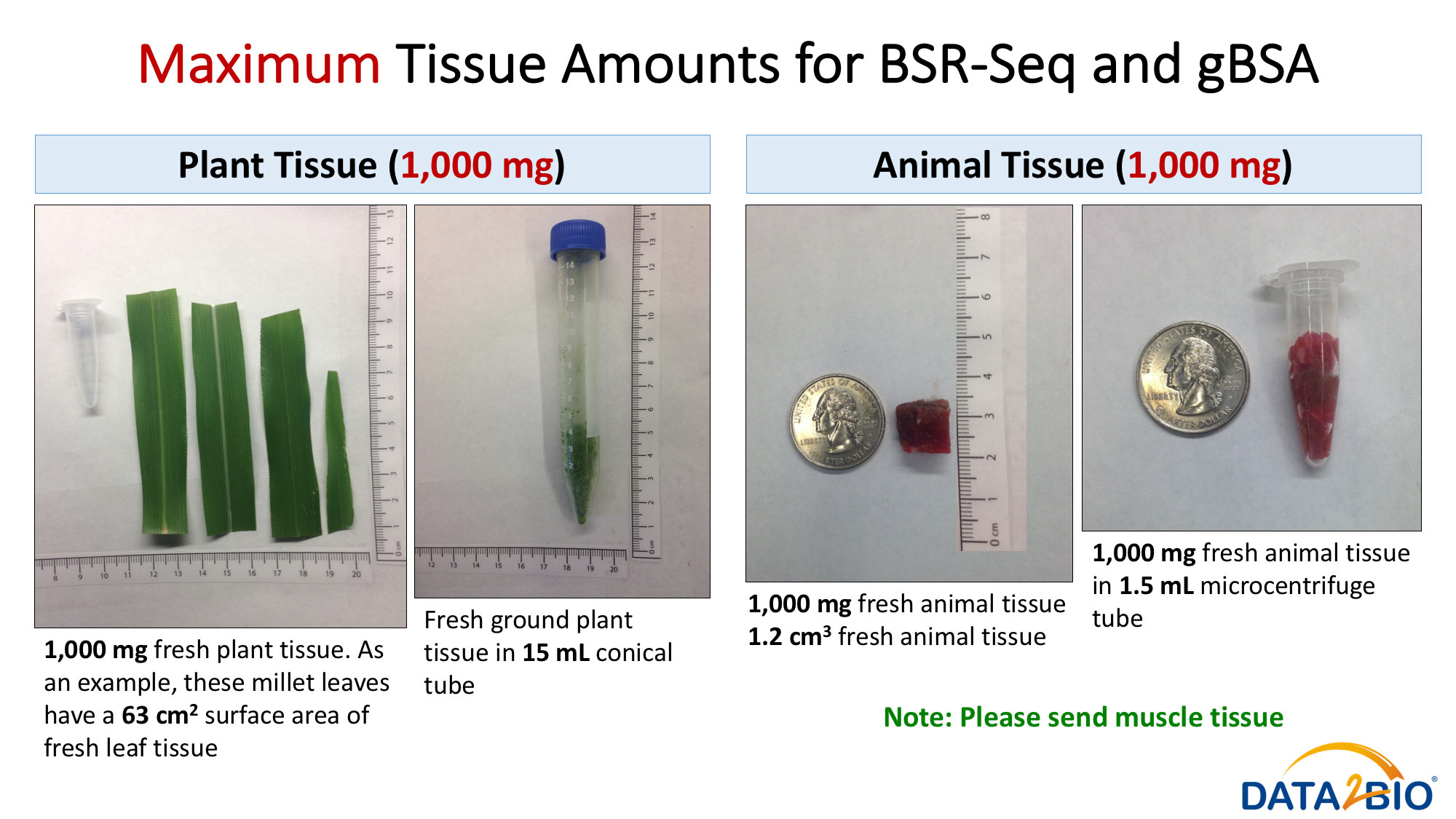
Please see pictures for details related to preferred and maximum tissue amounts required.
Preparing your samples for shipment
Preferred Method
Please send us your frozen DNA samples in EB buffer with dry ice in a Styrofoam container via overnight express. For samples submitted in 1.5 or 2.2 mL tubes, we strongly suggest either using screw capped tubes or sealing the lids on using parafilm. For samples submitted in 96 or 384-well plates, carefully seal each plate using sealing tape rather than foil and then wrap each plate individually. Use this template form to create your sample list and include a hard copy print out in the package. Seal the Styrofoam container with packing tape to slow the sublimation of the dry ice.
Alternative Method
Many of our customers live in countries where overnight service to the United States is unreliable or not available. If you are unsure whether your samples will remain frozen long enough to reach Data2Bio’s facility, we recommend mailing dried DNA samples instead. When using this method, first precipitate your samples into a pellet by adding ethanol, then dry the samples using a speed vacuum. Drying times will vary depending on the original volume of your samples. A significant quantity of DNA is lost during the drying and re-elution process, so if you use the dried DNA method we ask that you provide at least 20 µgrams of DNA per sample.
Shipping
When you are ready to mail your samples please send the tracking number and a copy of your sample list to:
Ship your samples to:
Data2Bio, LLC.
Attn: Lisa Coffey
2052 Roy J. Carver Co-Laboratory
1111 WOI Road
Ames, IA 50011-1085 USA
It is also important to time the shipment of your samples so they will arrive on a weekday rather than a Saturday, Sunday, or US Holiday. For an updated list of US holidays which may influence package delivery check this website: http://www.theholidayschedule.com/post-office-holidays.php
Additional Notes
Some projects rely on data provided by the client rather than (or in addition to) data generated by Data2Bio. Client-supplied data are expected to arrive at Data2Bio at project initiation (e.g., when samples are provided). Delays in supplying such data are likely to delay project completion.
Sample Submission for Tissue (to be used for DNA isolation)
Sample Requirements
We prefer that you send us lyophilized (i.e. freeze dried) tissue samples. If you do not have the equipment on site for freeze drying you can overnight at room temp fresh tissue samples to us and we can lyophilize them at our facility. If you will be submitting large numbers of tissue samples, cannot lyophilize them, and are not confident mailed samples will reach us within twenty-four hours please contact us at to discuss a number of other strategies which can be employed for tissue sampling. Please do not send us frozen tissue samples as it is much more difficult for us to grind and extract DNA from samples which were frozen without being dried.
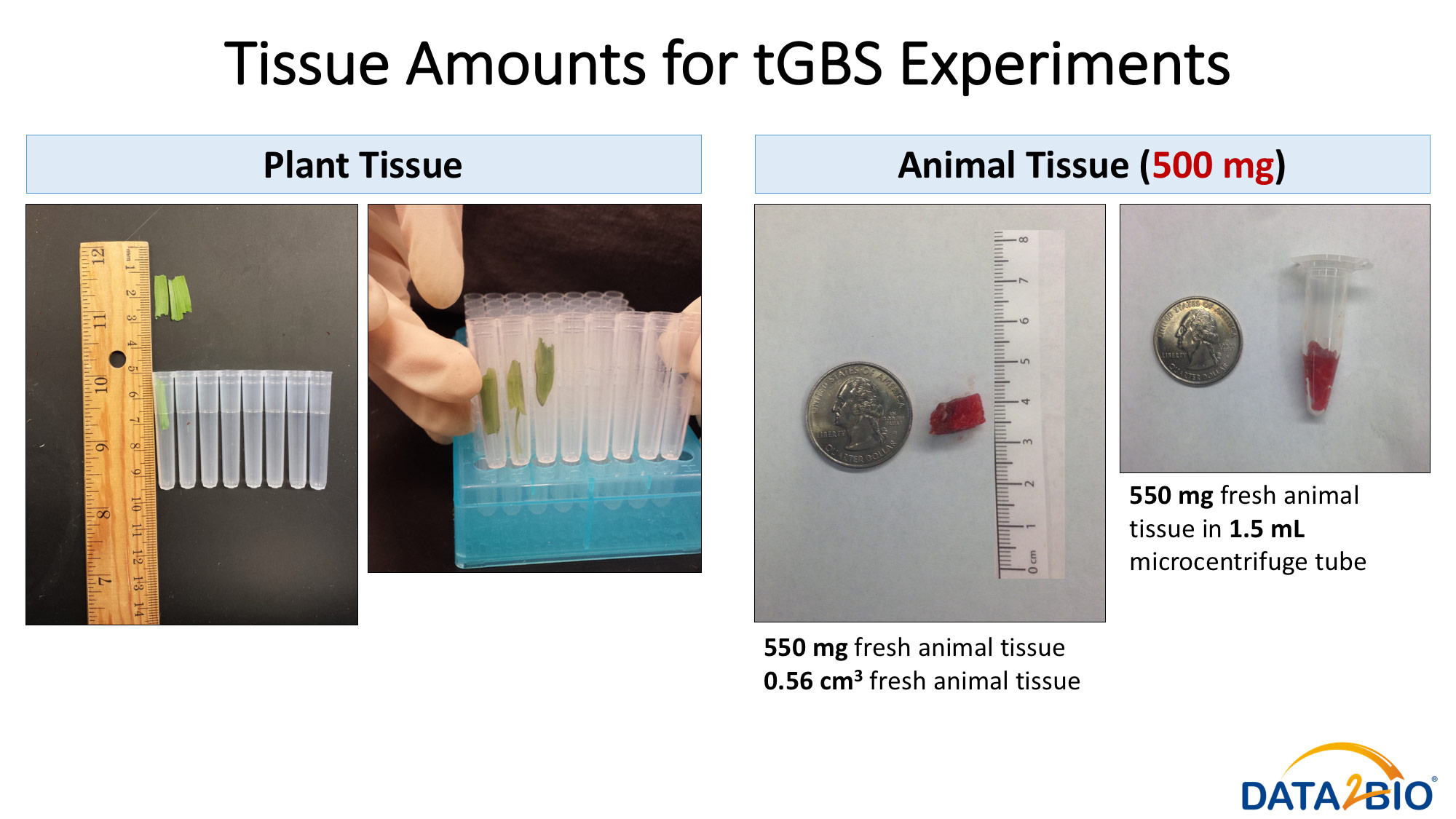
For tGBS® genotyping projects we ask that tissue samples be submitted in a 96 well format. For all projects, be sure the amount of tissue collected fits loosely in the container (96 well plate, 1.5 mL or 5 mL tube etc.). Tubes where too much tissue has been shoved into too small a space are also much more difficult to grind.
In addition, for tGBS® genotyping projects we ask for ~30 mg of freeze-dried (lyophilized) tissue per sample. We might be able to work with as little as 10 mg and samples should not be more than 100 mg of dried tissue per sample.
Supported Tissues
Plants
- We prefer young leaf tissue as they are both easy to grind and provide a high yield of purified DNA per unit of input tissue.
- We have also had success in extracting DNA from older leaves, stems, roots, pollen, fruits, roots, starchy tubers, and dried seeds. However please let us know if you will be submitting one of these tissues as it may influence our estimated completion date and/or quoted price.
Animals
- At this time we do not offer DNA extraction from animal tissues in house. We ask our customers performing genotyping in animal species to submit DNA samples.
Fungi
- Please contact us at
Shipping
When you are ready to mail your samples please send the tracking number and a copy of your sample list to:
Ship your samples to:
Data2Bio, LLC.
Attn: Lisa Coffey
2052 Roy J. Carver Co-Laboratory
1111 WOI Road
Ames, IA 50011-1085 USA
It is also important to time the shipment of your samples so they will arrive on a weekday rather than a Saturday, Sunday, or US Holiday. For an updated list of US holidays which may influence package delivery check this website: http://www.theholidayschedule.com/post-office-holidays.php
Additional Notes
Some projects rely on data provided by the client rather than (or in addition to) data generated by Data2Bio. Client-supplied data are expected to arrive at Data2Bio at project initiation (e.g., when samples are provided). Delays in supplying such data are likely to delay project completion.
Sample Submission for tGBS® Genotyping-by-Sequencing (GBS) Projects
If you will be submitting tissue samples for DNA isolation and tGBS® genotyping, please refer to our tissue sample submission FAQ and then return here for additional tGBS-specific information.
Sample Requirements
While the construction of tGBS® libraries is relatively insensitive to DNA quality, low quality samples can interfere with our normalization process and result in greater variance in the number of sequence reads generated per sample and the overall quality of the project. For this reason, we request that you send us DNA samples that have been extracted using Qiagen kits (DNeasy kit or Magattract Core kit) or equivalent. For best results, we recommend employing a commercial high throughput DNA isolation kit such as Qiagen's DNeasy or Magattract Core lines.
DNA quantification requirements1
- It is highly recommended that customers quantify their samples by fluorescent based methods (i.e. Qubit, PicoGreen Assay)
- Recommended QC values based on fluorescence:
- At least 5 ng/µL in 100 µL volume = 0.5 µg per sample
- Recommended QC values based on fluorescence:
- In cases where customers are unable to quantify by fluorescence, we do accept QC values based on absorbance (i.e. NanoDrop)
- Recommended QC values based on absorbance:
- At least 75 ng/µL in 100 µL volume = 7.5 µg per sample
- 260/280 ratio of each sample should be between 1.7-2.0
- Recommended QC values based on absorbance:
1 When necessary we can often work with smaller sample quantities. Please contact us at
What happens if some of my samples fail QC?
We will QC your samples upon their arrival, and let you know if any of them fall outside the expected range.
- If an entire plate of samples exhibits unacceptable quality (as is often seen with CTAB-extracted DNA) we may offer to re-purifying the entire plate at our facility which can often remove contaminants that influence DNA quality estimates and quantification. This will, however, incur an additional charge.
- For logistical reasons, we are not able swap out (or re-array) individual samples that have failed to pass QC.
- If samples do fail QC (e.g., because of insufficient DNA) and we cannot purify them, you are welcome to send an entire plate as a replacement. Please realize that this will delay of project completion while we wait for replacement samples (we won't proceed with the project until all samples have passed QC).
- If you prefer, you can also submit lyophilized tissue samples and Data2Bio will isolate and quantify DNA. This will, however, incur an additional DNA extraction charge.
Naming Samples for Data2Bio's tGBS® Analysis Pipeline
Each unique genotype submitted for tGBS® should have a unique ID consisting of only alpha-numeric characters and optionally underscores2. During analysis lowercase letters contained with sample identifiers will be converted to uppercase letters and all special characters will be replaced with underscores3. In addition, white space (blank) characters leading and/or trailing sample IDs will also be removed.
If you submit multiple plates of samples, each unique genotype should have an identifier that distinguishes not only from other genotypes on the same plate but across all submitted plates. In the event of projects requiring the use of the data generated from previous projects, please ensure that the new samples have unique identifiers relative to the previous project if samples are meant to be kept separate.
Our default analysis pipeline treats all samples having the same ID (regardless of case, within plate, or between multiple plates) as the same genotype and the generated reads from those sample wells will be pooled (combined) in downstream analyses. Sometimes clients intentionally replicate the same genotype among their samples. If you wish these replicates to be pooled during our post-sequencing analysis (this is our recommendation for parental controls) then you should use the identical sample ID for all the replicates (e.g., ABC, ABC etc.). If, on the other hand, you do not wish the replicates to be pooled during sequence analysis they should be given unique IDs (e.g., append an underscore followed by a number after the common ID shared by all replicates: ABC_1, ABC_2 etc).
2 We realize some sample IDs contain dashes. Our analysis pipeline will automatically replace the dashes with underscores. Please alert us if that will be a problem.
3 Please contact us if you have a special case or a more complicated situation.
Preparing your samples for shipment (Required Method)
- For tGBS® all samples should be submitted in 96 well plates. Samples submitted in individual tubes will incur additional handling fees.
- Please send all your samples at the same time. We will not proceed with library prep until all samples have arrived at our facility and have passed QC.
- Please send frozen DNA samples eluted in EB buffer or water.
- Do NOT send DNA eluted in solutions with high concentrations of EDTA (such as TE buffer).
- In some circumstances with highly concentrated DNA, we can genotype samples sent in TE buffer, but only if you inquire and get approval from our QC team before mailing your samples.
- Carefully seal each plate using sealing caps (either rubber mat or plastic caps) and wrap each plate individually
with bubble wrap before placing into Styrofoam container.
- Avoid foil because it be punctured during shipping.
- Avoid film because it can come loose from plate, especially if samples thaw.
- Pack Styrofoam container with dry ice
- Use this template form to create your sample list and include a hard copy print out in the package
- Seal the Styrofoam container with package tape to slow the sublimation of dry ice
- Ship via overnight express
- See further below about shipping
Some of our customers live in countries where overnight service to the United States is unreliable or not available. We have tested a number of protocols for shipping samples internationally, please contact us directly at to discuss which method is most suited to your situation.
Shipping
When you are ready to mail your samples please send the tracking number and a copy of your sample list to:
Ship your samples to:
Data2Bio, LLC.
Attn: Lisa Coffey
2052 Roy J. Carver Co-Laboratory
1111 WOI Road
Ames, IA 50011-1085 USA
It is also important to time the shipment of your samples so they will arrive on a weekday rather than a Saturday, Sunday, or US Holiday. For an updated list of US holidays which may influence package delivery check this website: http://www.theholidayschedule.com/post-office-holidays.php
Additional Notes
Some projects rely on data provided by the client rather than (or in addition to) data generated by Data2Bio. Client-supplied data are expected to arrive at Data2Bio at project initiation (e.g., when samples are provided). Delays in supplying such data are likely to delay project completion.